PRECIP EVAPOTRANS GR.WAT.FLOW RUNOFF STREAMFLOW -----------------(cm)---------------------------------- Apr 3.3 2.2 1.5 0.0 1.5 May 4.9 7.2 0.8 0.0 0.8 June 13.2 11.3 0.0 0.1 0.2 July 12.5 11.5 0.4 0.5 0.9 Aug 6.4 9.8 0.0 0.0 0.0 Sept 7.9 7.2 0.0 0.0 0.0 Oct 3.6 4.5 0.0 0.0 0.0 Nov 6.7 1.0 0.0 0.0 0.0 Dec 7.6 0.5 4.1 0.3 4.4 Jan 4.0 0.2 5.6 0.2 5.8 Feb 9.4 0.2 3.8 1.0 4.8 Mar 6.9 0.7 9.7 1.5 11.2 --------------------------------------------------------------- YEAR 86.4 56.4 26.0 3.6 29.6 Lets create function to obtain annual and monthly sums
I am learning R
About the Blog
Contributors
Archieves
Labels
- Basic (7)
- CRD (1)
- data access (1)
- data creation (1)
- data steps (3)
- gl (1)
- Introduction (5)
- packages (3)
- R (8)
- read.fwf (1)
- read.table (1)
- rep (1)
- scan (1)
- seq (1)
- sequence (1)
- xlsRedWrite (1)
Thursday, October 29, 2009
Multiple variables in the same scatter plot
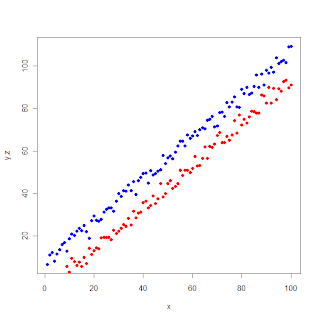
legend(10,85,legend = c("y","z"), col = c("blue", "red"),pch = 16)
# here 10, and 85 are the xy-coordinates where the legend-box will be added on the graph.
Now the graph is complete.
Saturday, February 21, 2009
Article about R in NYTimes
I was just browsing the net and found an interesting and very good article in NYtimes.
I am touched by the first sentence. R could be just 18th letter of English alphabet ………………., that’s exactly what happened many times when I mentioned R in some friends circle.
Wednesday, February 11, 2009
Data accessing, editing and Manipulation
lets create some vectors,
1: Var1<-c(1,2,3,1,2,4,5,0,2,6,8,5,3,7,5,2,8,9,2,10) 2: Var2<-rep(c(1,2,3,4) each=5) 3: Var3<-rep(1:5, 4)now, lets make a data frame
1: > datalist<-data.frame(Var1, Var2, Var3)
Var1 Var2 Var3
1 1 1 1
2 2 1 2
3 3 1 3
4 1 1 4
5 2 1 5
6 4 2 1
7 5 2 2
8 0 2 3
9 2 2 4
10 6 2 5
11 8 3 1
12 5 3 2
13 3 3 3
14 7 3 4
15 5 3 5
16 2 4 1
17 8 4 2
18 9 4 3
19 2 4 4
20 10 4 5
so, the variable names can be changed now with,
> names(datalist)<-c("X", "Y", "Z")
Data Access:
> datalist[1:4,]
X Y Z
1 1 1 1
2 2 1 2
3 3 1 3
4 1 1 4
> datalist[1:3,1:3]
X Y Z
1 1 1 1
2 2 1 2
3 3 1 3
> datalist[3,3]
[1] 3
> datalist$X[c(2,5,8)]
[1] 2 2 0
> datalist$X[-c(2,5,8)]
[1] 1 3 1 4 5 2 6 8 5 3 7 5 2 8 9 2 10
> datalist[c(2,5,8),c(1,2)]
X Y
2 2 1
5 2 1
8 0 2
Note that, X variable can not be accessed directly and we used datalist$X , if we want to access X directly we can use attach() function
> X
Error: object "X" not found
> attach(datalist)
> X
[1] 1 2 3 1 2 4 5 0 2 6 8 5 3 7 5 2 8 9 2 10
Tuesday, February 10, 2009
Generating treatment maps for a typical CRD experiment
Hellooo!
Saturday, February 7, 2009
Data Steps/ Reading from file
we seldom create the data our selves, most of the time we read the data from file. there are many ways to read the data.
Reading from Raw data
> abc<-read.table("control_vs_inocul.txt")
> abc[1:10,]V1 V2 V3 V4 V5
1 Rep Genotype top root Control
2 1 R1R4 0.65 0.48 0
3 1 R1R7 0.77 0.32 0
4 1 R1M1 0.77 0.25 0
5 1 R1M3 0.73 0.31 0
6 1 R1S1 0.69 0.33 0
7 1 R1S3 0.72 0.34 0
8 1 R1S7 0.56 0.28 0
9 1 R4R7 0.69 0.37 0
10 1 R4M1 0.65 0.40 0
Notice the V1, V2....... these are the default variables given by the R system , so to inclue the variable name from file
> abc<-read.table("control_vs_inocul.txt", header=T)
> abc[1:10,]Rep Genotype top root Control
1 1 R1R4 0.65 0.48 0
2 1 R1R7 0.77 0.32 0
3 1 R1M1 0.77 0.25 0
4 1 R1M3 0.73 0.31 0
5 1 R1S1 0.69 0.33 0
6 1 R1S3 0.72 0.34 0
7 1 R1S7 0.56 0.28 0
8 1 R4R7 0.69 0.37 0
9 1 R4M1 0.65 0.40 0
10 1 R4M3 0.86 0.30 0
or to read the data first and assign variable name later,
> abc<-read.table("control_vs_inocul.txt", header=F, skip=1)
> abc[1:10,]V1 V2 V3 V4 V5
1 1 R1R4 0.65 0.48 0
2 1 R1R7 0.77 0.32 0
3 1 R1M1 0.77 0.25 0
4 1 R1M3 0.73 0.31 0
5 1 R1S1 0.69 0.33 0
6 1 R1S3 0.72 0.34 0
7 1 R1S7 0.56 0.28 0
8 1 R4R7 0.69 0.37 0
9 1 R4M1 0.65 0.40 0
10 1 R4M3 0.86 0.30 0
> names(abc)<-c("A", "B", "C", "D", "E")
> abc[1:10,]A B C D E
1 1 R1R4 0.65 0.48 0
2 1 R1R7 0.77 0.32 0
3 1 R1M1 0.77 0.25 0
4 1 R1M3 0.73 0.31 0
5 1 R1S1 0.69 0.33 0
6 1 R1S3 0.72 0.34 0
7 1 R1S7 0.56 0.28 0
8 1 R4R7 0.69 0.37 0
9 1 R4M1 0.65 0.40 0
10 1 R4M3 0.86 0.30 0
in some rows there are missing data..
> abc[210:215,]A B C D E
210 3 R7M1 0.60 0.26 1
211 3 R7M3 0.36 0.31 1
212 3 R7S1 0.68 0.23 1
213 3 R7S3 . . 1
214 3 R7S7 0.52 0.42 1
215 3 M1M3 0.58 0.13 1
here "." does not mean missing value. it is the input from file, so to recognize it as missing value
> abc<-read.table("control_vs_inocul.txt", header=T, na.string=".")
> abc[210:215,]Rep Genotype top root Control
210 3 R7M1 0.60 0.26 1
211 3 R7M3 0.36 0.31 1
212 3 R7S1 0.68 0.23 1
213 3 R7S3 NA NA 1
214 3 R7S7 0.52 0.42 1
215 3 M1M3 0.58 0.13 1
the default separator of read.table is " ", you can always change this in parameter sep="," or any other else.
for a complete description type ?read.table in R interface.
there are variations in read.table such as read.csv, read.delim
The another common format of raw data is fixed width format. to read that you can use read.fwf
for example if fwf1.dat contains following data
1S1.52.33
2S2.56.33
3R1.23
then
> def <- read.fwf("fwf1.dat", width=c(1,1,3,4), col.names=c("ID", "type", "top", "root"))
> def
ID type top root
1 1 S 1.5 2.33
2 2 S 2.5 6.33
3 3 R 1.2 3.00
Another primitive function to read data is scan() function
> scanned<-scan("control_vs_inocul.txt", skip=1, what=list(0,"",0,0,0), nlines=7)
Read 7 records
> scanned
[[1]]
[1] 1 1 1 1 1 1 1
[[2]]
[1] "R1R4" "R1R7" "R1M1" "R1M3" "R1S1" "R1S3" "R1S7"
[[3]]
[1] 0.65 0.77 0.77 0.73 0.69 0.72 0.56
[[4]]
[1] 0.48 0.32 0.25 0.31 0.33 0.34 0.28
[[5]]
[1] 0 0 0 0 0 0 0
you can also use scan to directly input data with keyboard.
> scan()
1: 1 2 3 4 5 6 7 8
9:
Read 8 items
[1] 1 2 3 4 5 6 7 8
Most of the time we input our raw data in spreadsheet application such as MS Excel. What I do is save the data in csv or tab delimitated txt format and read through read.table function.
Another nice package to read data directly from Excel is xlsReadWrite. to import from Excel file directly,
> library(xlsReadWrite)
> data1<-read.xls("original.xls")> data1[1:7,]
F M rep. CROSS Egg Gall top root. count
1 1 2 1 12 0 1 0.65 0.48 1371
2 1 2 1 12 0 1 0.65 0.48 1371
3 1 2 1 12 1 1 0.65 0.48 1371
4 1 2 1 12 0 1 0.65 0.48 1371
5 1 2 1 12 1 3 0.65 0.48 1371
6 1 2 1 12 0 1 0.65 0.48 1371
7 1 2 1 12 0 1 0.65 0.48 1371
for more information about the parameters and default values look at
http://cran.r-project.org/web/packages/xlsReadWrite/xlsReadWrite.pdf
further next time
Sunday, July 27, 2008


